Overview
Neural decoding is a data analysis method that uses pattern classifiers to predict experimental conditions based on neural activity. The NeuroDecodeR package makes it easy to do neural decoding analyses in R.
Installation
You can install NeuroDecodeR package from CRAN of the development version on GitHub.
# Current release on CRAN:
install.packages("NeuroDecodeR")
# Development version on GitHub:
# install.packages("devtools")
devtools::install_github("emeyers/NeuroDecodeR")Documentation
The documentation for this package is available at: https://emeyers.github.io/NeuroDecodeR/
To get started we recommend you read the introductory tutorial
Usage
The package is based on 5 abstract object types:
-
Datasources (DS): generate training and test sets. -
Feature preprocessors (FP): apply preprocessing to the training and test sets. -
Classifiers (CL): learn relationships on the training set and make predictions on the test data. -
Result Metrics (RM): summarize the prediction accuracies. -
Cross-validators (CV): take the DS, FP and CL objects and run a cross-validation decoding procedure.
By combing different versions of these 5 object types together, it is possible to run a range of different decoding analyses.
Below is a brief illustration of how to use the NDR to do a simple decoding analysis. To learn how to use the NDR please see the documentation website and the package vignettes.
library(NeuroDecodeR)
# file to data in "binned format"
basedir_file_name <- system.file(file.path("extdata", "ZD_150bins_50sampled.Rda"),
package="NeuroDecodeR")
# create the DS, FP, CL, RM, and CV objects
ds <- ds_basic(basedir_file_name, 'stimulus_ID', 5, num_label_repeats_per_cv_split = 3)
#> Automatically selecting sites_IDs_to_use. Since num_cv_splits = 5 and num_label_repeats_per_cv_split = 3, all sites that have 15 repetitions have been selected. This yields 132 sites that will be used for decoding (out of 132 total).
fps <- list(fp_zscore())
cl <- cl_max_correlation()
rms <- list(rm_main_results(), rm_confusion_matrix())
cv <- cv_standard(datasource = ds,
classifier = cl,
feature_preprocessors = fps,
result_metrics = rms,
num_resample_runs = 2) # better to use more resample runs for actual analyses (default is 50)
# run a decoding analysis (this takes a few minutes)
DECODING_RESULTS <- run_decoding(cv)
#> | | | 0% | |=================================== | 50% | |======================================================================| 100%
# plot the results for three different result types
plot(DECODING_RESULTS$rm_main_results, results_to_show = 'all', type = 'line')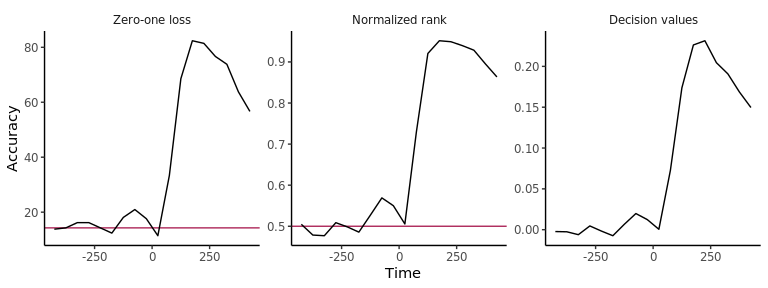
# create a temporal cross decoding plot
plot(DECODING_RESULTS$rm_main_results)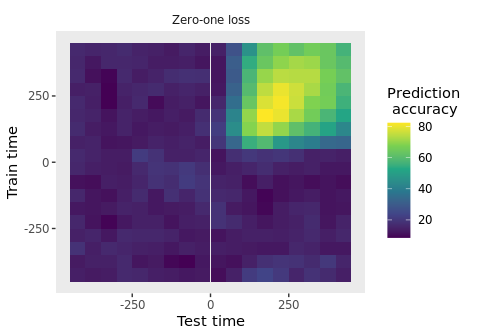
Running an analysis using pipes (|>)
One can also run a decoding analysis using the pipe (|>) operator to string together the different NDR objects as shown below.
basedir_file_name <- system.file(file.path("extdata", "ZD_500bins_500sampled.Rda"), package="NeuroDecodeR")
DECODING_RESULTS <- basedir_file_name |>
ds_basic('stimulus_ID', 6, num_label_repeats_per_cv_split = 3) |>
cl_max_correlation() |>
fp_zscore() |>
rm_main_results() |>
rm_confusion_matrix() |>
cv_standard(num_resample_runs = 2) |>
run_decoding()
#> Automatically selecting sites_IDs_to_use. Since num_cv_splits = 6 and num_label_repeats_per_cv_split = 3, all sites that have 18 repetitions have been selected. This yields 132 sites that will be used for decoding (out of 132 total).
#> | | | 0% | |=================================== | 50% | |======================================================================| 100%
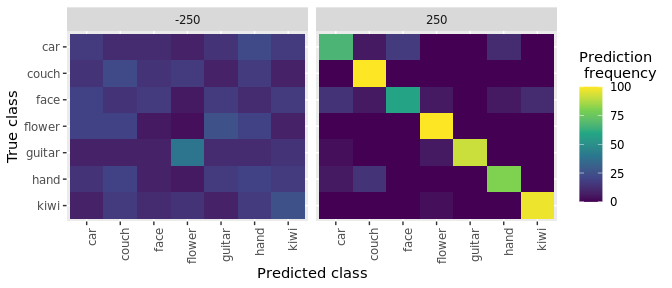
plot(DECODING_RESULTS$rm_confusion_matrix)