Overview
Neural decoding is a data analysis method that uses pattern classifiers to predict experimental conditions based on neural activity. The Neural Decoding Toolbox in R (NDTr) makes it easy to do neural decoding analyses in R.
Usage
The package is based on 5 abstract object types:
-
Datasources (DS): generate training and test sets. -
Feature preprocessors (FP): apply preprocessing to the training and test sets. -
Classifiers (CL): learn relationships on the training set and make predictions on the test data. -
Result Metrics (RM): summarize the prediction accuracies. -
Cross-validators (CV): take the DS, FP and CL objects and run a cross-validation decoding procedure.
By combing different versions of these 5 object types together, it is possible to run a range of different decoding analyses.
Below is a brief illustration of how to use the NDTr to do a simple decoding analysis. To learn how to use the NDTr please see the documentation website and the package vignettes.
library(NDTr)
# file to data in "binned format"
basedir_file_name <- system.file(file.path("extdata", "ZD_150bins_50sampled.Rda"), package="NDTr")
# create the DS, FP, CL, RM, and CV objects
ds <- ds_basic(basedir_file_name, 'stimulus_ID', 5, num_label_repeats_per_cv_split = 3)
fps <- list(fp_zscore())
cl <- cl_max_correlation()
rms <- list(rm_main_results(aggregate_normalized_rank = "diag"), rm_confusion_matrix())
cv <- cv_standard(ds, cl, fps, rms, 3)
# run a decoding analysis (this takes a few minutes)
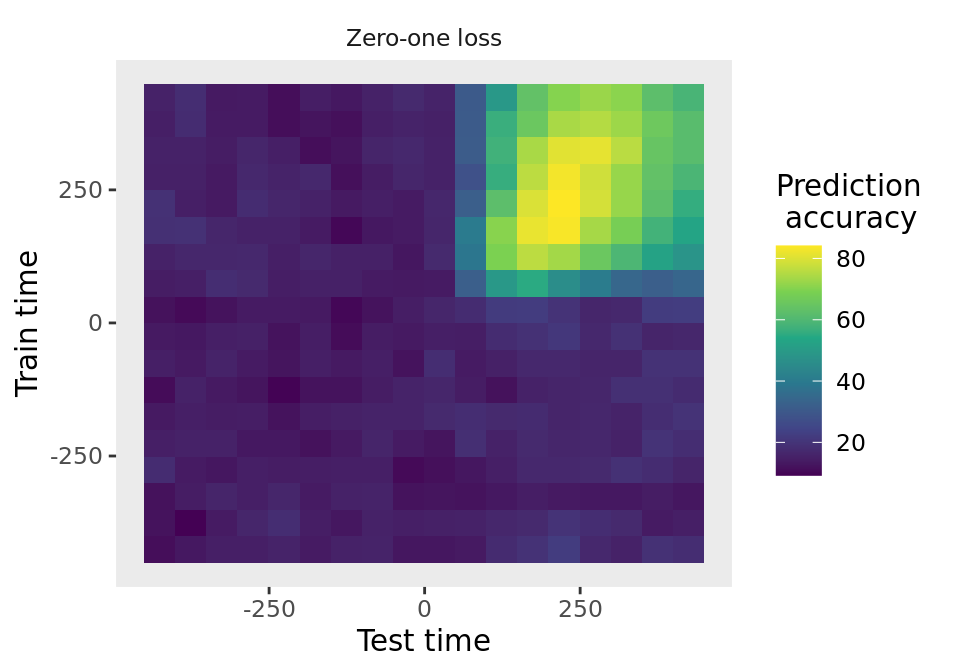
DECODING_RESULTS <- run_decoding(cv)# plot the results for three different result types
plot(DECODING_RESULTS$rm_main_results, result_type = "all", plot_type = "line")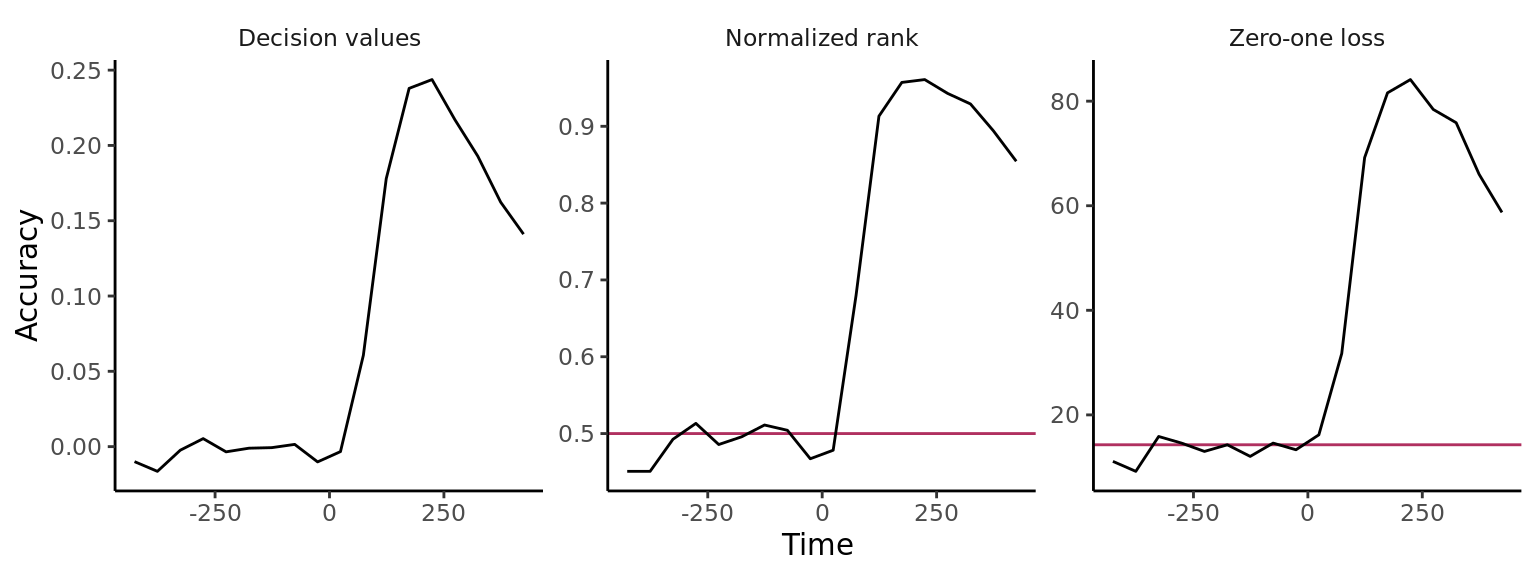
Documentation
The documentation for this package is available at: https://emeyers.github.io/NDTr/
To get started we recommend you read the introductory tutorial